c3s_sm¶
Reading and reshuffling of C3S soil moisture Written in Python.
Installation¶
Setup of a complete environment with conda can be performed using the following commands:
git clone git@github.com:TUW-GEO/c3s_sm.git c3s_sm
cd c3s_sm
conda env create -f environment.yml
source activate c3s_sm
Supported Products¶
At the moment this package supports C3S soil moisture data in netCDF format (reading and time series creation) with a spatial sampling of 0.25 degrees.
Contribute¶
We are happy if you want to contribute. Please raise an issue explaining what is missing or if you find a bug. We will also gladly accept pull requests against our master branch for new features or bug fixes.
Development setup¶
For Development we also recommend a conda environment. You can create one
including test dependencies and debugger by running
conda env create -f environment.yml. This will create a new c3s_sm
environment which you can activate by using source activate c3s_sm.
Guidelines¶
If you want to contribute please follow these steps:
- Fork the c3s_sm repository to your account
- Clone the repository, make sure you use
git clone --recursiveto also get the test data repository. - make a new feature branch from the c3s_sm master branch
- Add your feature
- Please include tests for your contributions in one of the test directories. We use py.test so a simple function called test_my_feature is enough
- submit a pull request to our master branch
Note¶
This project has been set up using PyScaffold 2.5. For details and usage information on PyScaffold see http://pyscaffold.readthedocs.org/.
Reading C3S SM images¶
Reading of the C3S SM raw netcdf files can be done in two ways.
Reading by file name¶
import os
from datetime import datetime
from c3s_sm.interface import C3SImg
import numpy.testing as nptest
# read several parameters
parameter = ['sm', 'sm_uncertainty']
# the class is initialized with the exact filename.
image_path = os.path.join(os.path.dirname(__file__), 'tests', 'c3s_sm-test-data',
'img', 'ICDR', '060_dailyImages', 'combined', '2017')
image_file = 'C3S-SOILMOISTURE-L3S-SSMV-COMBINED-DAILY-20170701000000-ICDR-v201706.0.0.nc'
img = C3SImg(os.path.join(image_path, image_file), parameter=parameter)
# reading returns an image object which contains a data dictionary
# with one array per parameter. The returned data is a global 0.25 degree
# image/array.
image = img.read()
assert image.data['sm'].shape == (720, 1440)
assert image.lon.shape == (720, 1440)
assert image.lon.shape == image.lat.shape
assert image.lon[0, 0] == -179.875
assert image.lat[0, 0] == 89.875
assert sorted(image.data.keys()) == sorted(parameter)
assert(image.metadata['sm']['long_name'] == 'Volumetric Soil Moisture')
nptest.assert_almost_equal(image.data['sm'][167, 785], 0.14548, 4)
Reading by date¶
All the C3S SM data in a directory structure can be accessed by date. The filename is automatically built from the given date.
from c3s_sm.interface import C3S_Nc_Img_Stack
parameter = 'sm'
img = C3S_Nc_Img_Stack(data_path=os.path.join(os.path.dirname(__file__),
'tests', 'c3s_sm-test-data', 'img',
'ICDR', '061_monthlyImages', 'passive'),
parameter=parameter)
image = img.read(datetime(2017, 7, 1, 0))
nptest.assert_almost_equal(image.data['sm'][167, 785], 0.23400, 4)
For reading all image between two dates the
c3s_sm.interface.C3S_Nc_Img_Stack.iter_images() iterator can be
used.
Conversion to time series format¶
For a lot of applications it is favorable to convert the image based format into a format which is optimized for fast time series retrieval. This is what we often need for e.g. validation studies. This can be done by stacking the images into a netCDF file and choosing the correct chunk sizes or a lot of other methods. We have chosen to do it in the following way:
Store only the reduced gaußian grid points since that saves space.
Further reduction the amount of stored data by saving only land points if selected.
Store the time series in netCDF4 in the Climate and Forecast convention Orthogonal multidimensional array representation
Store the time series in 5x5 degree cells. This means there will be 2566 cell files (1001 when reduced to land points) and a file called
grid.ncwhich contains the information about which grid point is stored in which file. This allows us to read a whole 5x5 degree area into memory and iterate over the time series quickly.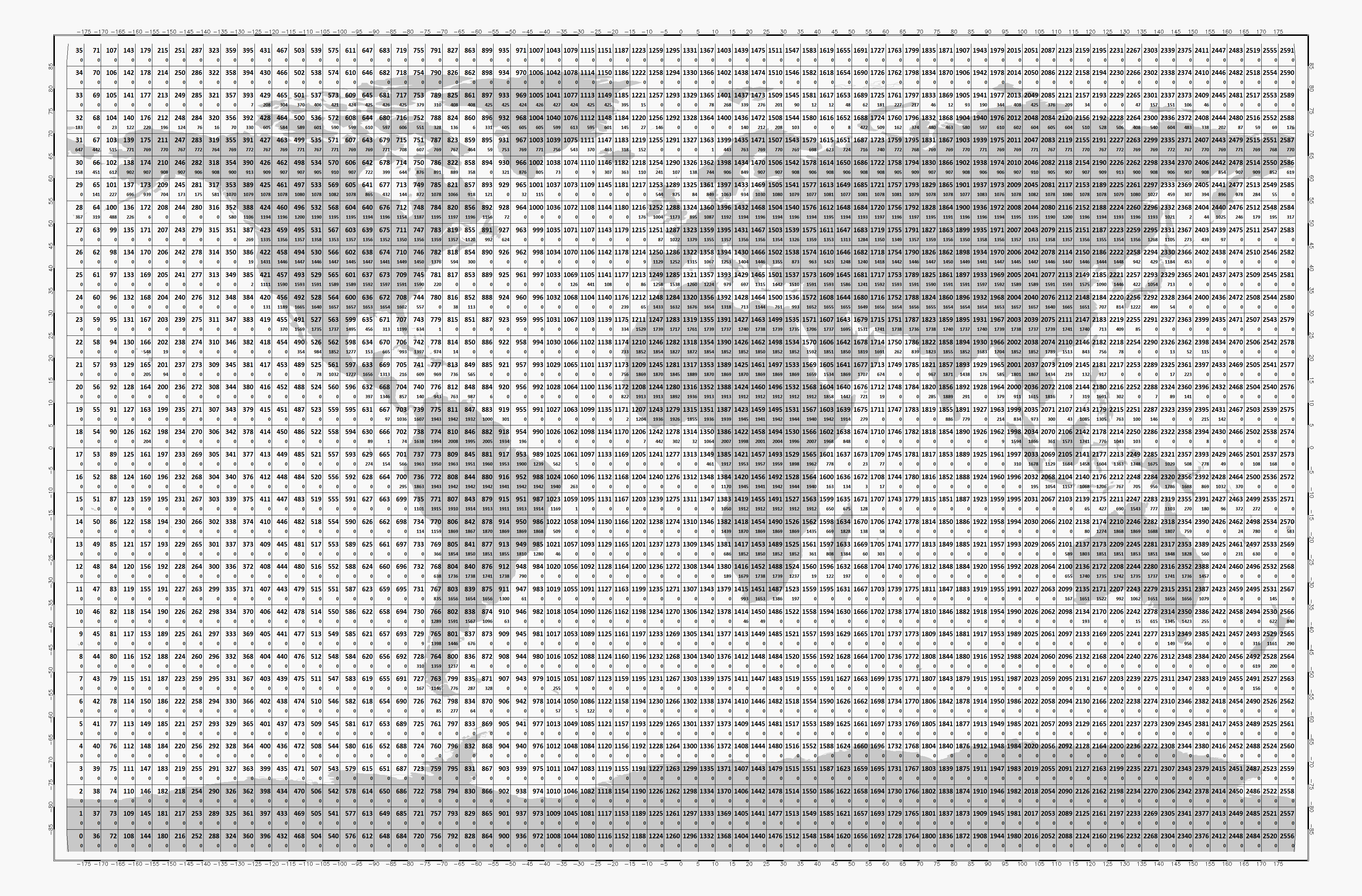
This conversion can be performed using the c3s_repurpose command line
program. An example would be:
c3s_repurpose /c3s_images /timeseries/data 2000-01-01 2001-01-01 sm sm_uncertainty --land_points True
Which would take C3S SM data stored in /c3s_images from January 1st
2000 to January 1st 2001 and store the parameters for soil moisture and its uncertainty
of points marked as ‘land’ in the smecv-grid as time
series in the folder /timeseries/data.
Note: If a RuntimeError: NetCDF: Bad chunk sizes. appears during reshuffling, consider downgrading the
netcdf4 C-library via:
conda install -c conda-forge libnetcdf==4.3.3.1 --yes
Conversion to time series is performed by the repurpose package in the background. For custom settings
or other options see the repurpose documentation and the code in
c3s_sm.reshuffle.
Reading converted time series data¶
For reading the data the c3s_repurpose command produces the class
C3STs can be used:
from c3s_sm.interface import C3STs
ds = C3STs(ts_path)
# read_ts takes either lon, lat coordinates or a grid point indices.
# and returns a pandas.DataFrame
ts = ds.read_ts(45, 15)
Variable names for C3S Soil Moisture¶
C3S SM variables as in the netcdf image files (and time series from netcdf images) for different products and versions of C3S
- Combined Product
| short_name | Parameter | Units |
|---|---|---|
| freqbandID | Frequency Band Identification | |
| lat | Latitude | [degrees_north] |
| lon | Longitude | [degrees_east] |
| nobs | Number of valid observation | |
| sensor | Sensor Flag | |
| sm | Volumetric Soil Moisture | [m3 m-3] |
| time | Time | [days since 1970-01-01 00:00:00 UTC] |